Abstract
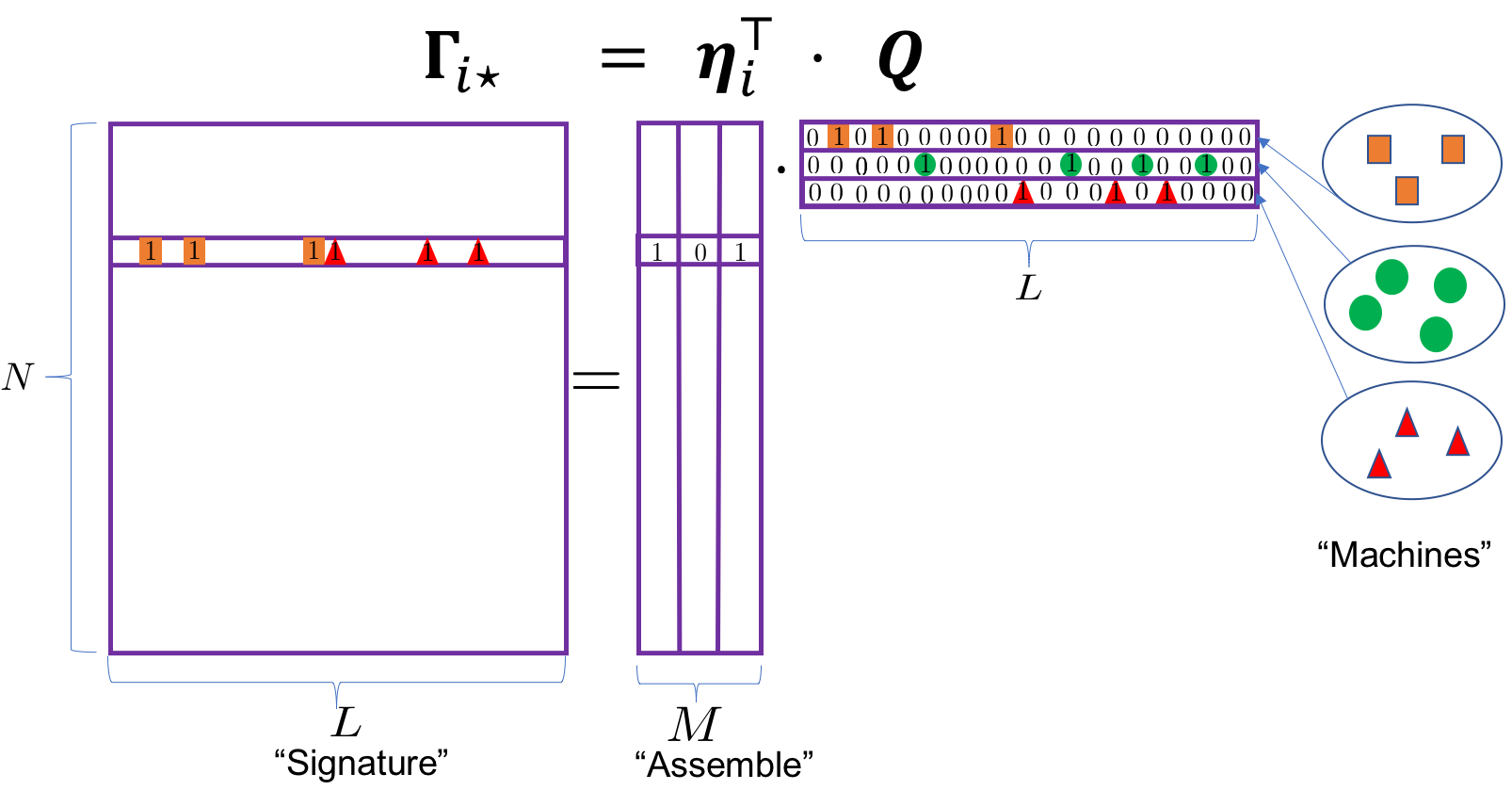
An ongoing challenge in subsetting autoimmune disease patients is how to summarize autoantibody mixture in terms of reactions to elemental sets of autoantigens. In this paper, we develop a model to support this type of substantive research. Our approach is to posit a mixed membership regression model for discovering autoantibody response profiles among patients, in which the mixing weights are parametrized by observed covariates. In this model, the profiles are unobserved multivariate binary presence or absence status measured with error and the profile prevalence is specified as a parsimonious stick-breaking regression model on an arbitrary number of patient-level covariates, such as cancer type, age and cancer-scleroderma interval, enabling clinicians to introduce elements of phenotypes related to the autoantibody profiles into the model. We demonstrate the proposed method by analyzing patients’ gel electrophoresis autoradiography (GEA) data. Our method quantifies the variation of both the frequency and nature of autoantibody profiles by covariates.
Keywords: Autoimmune disease; Clustering; Dependent Binary Data; Latent Class Models; Markov Chain Monte Carlo; Measurement Error; Mixture of Finite Mixture Models; Scleroderma.
R Code here