Abstract
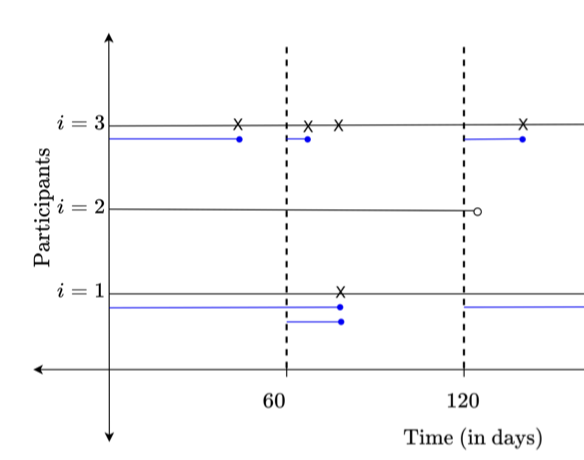
Recurrent events are common in clinical, healthcare, social and behavioral studies, yet methods for dynamic risk prediction of these events are limited. To overcome some long-standing challenges in analyzing censored recurrent event data, a recent regression analysis framework constructs a censored longitudinal data set consisting of times to the first recurrent event in multiple pre-specified follow-up windows of length τ (Xia, Murray and Tayob, 2020, XMT). Traditional regression models struggle with nonlinear and multi-way interactions, with success depending on the skill of the statistical programmer. With a staggering number of potential predictors being generated from genetic, -omic, and electronic health records sources, machine learning approaches such as the random forest regression are growing in popularity, as they can nonparametrically incorporate information from many predictors with nonlinear and multi-way interactions involved in prediction. In this paper, we (1) develop a random forest approach for dynamically predicting probabilities of remaining event-free during a subsequent τ-duration follow-up period from a reconstructed censored longitudinal data set, (2) modify the XMT regression approach to predict these same probabilities, subject to the limitations that traditional regression models typically have, and (3) demonstrate how to incorporate patient-specific history of recurrent events for prediction in settings where this information may be partially missing. We show the increased ability of our random forest algorithm for predicting the probability of remaining event-free over a τ-duration follow-up window when compared to our modified XMT method for prediction in settings where association between predictors and recurrent event outcomes is complex in nature. We also show the importance of incorporating past recurrent event history in prediction algorithms when event times are correlated within a subject. The proposed random forest algorithm is demonstrated using recurrent exacerbation data from the trial of Azithromycin for the Prevention of Exacerbations of Chronic Obstructive Pulmonary Disease (Albert and others, 2011).
Keywords Censored data; Longitudinal data; Pseudo-observations; Random forest; Recurrent events
Media Release: SPH News