Highlights:
-
Meat may be an important vehicle for human exposure to extraintestinal pathogenic E. coli strains from food animals
-
We present a novel approach for predicting the origins of clinical E. coli isolates
-
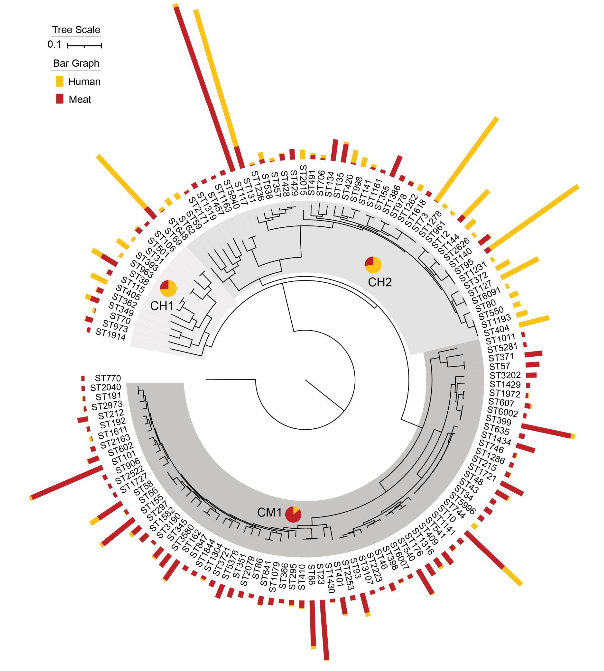
Approximately 8% of the clinical E. coli isolates in our population appeared to be foodborne zoonotic strains
-
Foodborne zoonotic E. coli strains were associated with asymptomatic bacteriuria, urinary tract infections, and sepsis
-
We estimate that foodborne zoonotic E. coli strains cause >480,000 urinary tract infections in the U.S. annually
Abstract
A one-health perspective may provide new and actionable information about Escherichia coli transmission. E. coli colonizes a broad range of vertebrates, including humans and food-production animals, and is a leading cause of bladder, kidney, and bloodstream infections in humans. Substantial evidence supports foodborne transmission of pathogenic E. coli strains from food animals to humans. However, the relative contribution of foodborne zoonotic E. coli (FZEC) to the human extraintestinal disease burden and the distinguishing characteristics of such strains remain undefined. Using a comparative genomic analysis of a large collection of contemporaneous, geographically-matched clinical and meat-source E. coli isolates (n = 3,111), we identified 17 source-associated mobile genetic elements – predominantly plasmids and bacteriophages – and integrated them into a novel Bayesian latent class model to predict the origins of clinical E. coli isolates. We estimated that approximately eight percent of human extraintestinal E. coli infections (mostly urinary tract infections) in our study population were caused by FZEC. FZEC strains were equally likely to cause symptomatic disease as non-FZEC strains. Two FZEC lineages, ST131-H22 and ST58, appeared to have particularly high virulence potential. Our findings imply that FZEC strains collectively cause more urinary tract infections than does any single non-E. coli uropathogenic species (e.g., Klebsiella pneumoniae). Our novel approach can be applied in other settings to identify the highest-risk FZEC strains, determine their sources, and inform new one-health strategies to decrease the heavy public health burden imposed by extraintestinal E. coli infections.
Keywords
- Escherichia coli;
- Infectious diseases;
- Urinary tract infection;
- Zoonosis;
- Foodborne;
- Mobile genetic element;
- Host adaptation;
- Food-animal production;
- Bayesian latent class model;
- Public health